Why validating iEEG biomarker detectors is so difficult
Interictal biomarkers of epilepsy such as spikes and high-frequency oscillations (HFOs) are increasingly being studied as clinical tools to guide epilepsy surgery. Since manually identifying these biomarkers in electrophysiological recordings is too time-consuming for standard clinical practice, several automated detectors have been introduced over the past decade. Yet, despite an active research field and important progress in algorithmic design, a fundamental issue remains: how should we validate detectors?
The issue might seem trivial at first. Ideally, one would run their novel detector on an annotated and independent testing dataset, calculate performance metrics by comparing detections with ground truth human annotations, and use obtained metrics to compare models across studies.
Challenge 1: Lack of standardized annotated datasets
The first major obstacle is the absence of high-quality benchmark datasets. Creating such resources is tedious and expensive, as it requires several experts to review long iEEG recordings, and manually label individual events. These events can be difficult to differentiate from background activity and artefacts. As a result, many studies rely on internally acquired data from small cohorts of patients, reviewed by local experts. These annotated datasets are not always public, and even when they are, their quality and generalizability are difficult to assess. Epilepsy centres differ substantially regarding acquisition equipment and protocols, sampling rates, filtering parameters, file formats, and annotation practices. Given this lack of standardization, validating a detector’s performance on data from a single centre is an unreliable benchmark. Collaborative datasets bringing together data from different centres, organized under shared annotation and formatting standards, are essential for both scientific progress and consequent patient care.
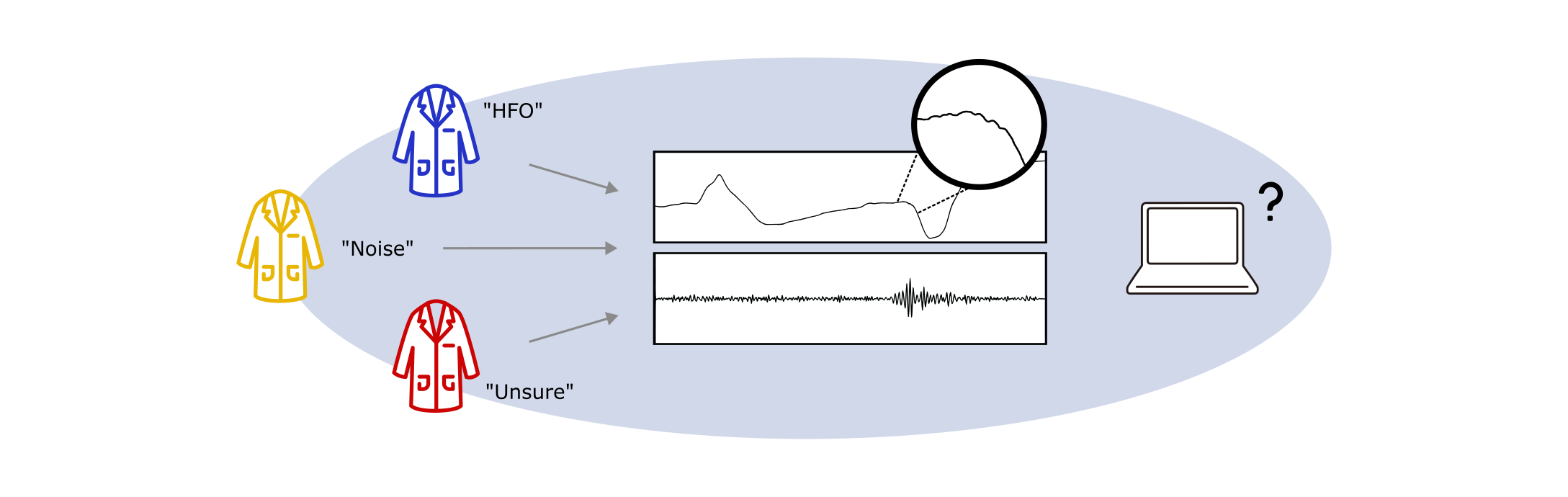
Challenge 2: What is the ground truth?
The idea of ‘standardized annotations’ implies an expert consensus on what constitutes an interictal biomarker. However, while epileptic spikes now have relatively recent EEG-based criteria defining what qualifies as a spike (Nascimento & Beniczky, 2023), intracranial-specific definitions are still lacking, and HFOs remain without formal consensus criteria. For both types of biomarkers, expert raters themselves frequently disagree on what constitutes an event (see for example Spring et al., 2017 or Scheuer et al., 2017). This disagreement introduces variability into testing datasets and makes it difficult to accurately measure a detector’s performance and compare across studies. A detector may in fact outperform a human reviewer but appear to underperform if the reviewer’s criteria for identifying HFOs are inaccurate or inconsistent.
What is the ground truth? When experts disagree on event annotations, establishing an objective ground truth against which to benchmark algorithms is a challenging task.
Challenge 3: Inconsistent validation protocols
Currently, studies differ in their evaluation procedures: different data sets, but also different performance metrics and testing conditions. Some report sensitivity and specificity, while others use accuracy, area under the receiver operating characteristic curve, or F1-score. This variability makes it difficult to benchmark new algorithms based on published metrics. While re-implementing existing algorithms on new datasets is possible, it requires access to up-to-date algorithm code and exhaustive documentation of testing procedures, such as data pre-processing steps, algorithm parameters, and whether the models are tested on continuous or event-segmented data. Such information is, however, often incomplete.
Going forward, the field needs to define essential standards for biomarker definitions, annotation guidelines, benchmark datasets and evaluation methodologies. Frameworks like SzCORE for seizure detection could be used as models (Pale et al., 2025). Building consensus in these areas will be essential to overcome validation challenges and move iEEG biomarker detection from research setting to clinical routine.
Authors: Clarissa Baratin, Emmanuel Barbeau - first published January 7th, 2026
References
Dan, J., Pale, U., Amirshahi, A., Cappelletti, W., Ingolfsson, T. M., Wang, X., ... & Ryvlin, P. (2025). SzCORE: seizure community open‐source research evaluation framework for the validation of electroencephalography‐based automated seizure detection algorithms. Epilepsia, 66, 14-24.
Nascimento, F. A., & Beniczky, S. (2023). Teaching the 6 criteria of the International Federation of Clinical Neurophysiology for defining interictal epileptiform discharges on EEG using a visual graphic. Neurology® Education, 2(2), e200073.
Scheuer, M. L., Bagic, A., & Wilson, S. B. (2017). Spike detection: Inter-reader agreement and a statistical Turing test on a large data set. Clinical Neurophysiology, 128(1), 243-250.
Spring, A. M., Pittman, D. J., Aghakhani, Y., Jirsch, J., Pillay, N., Bello-Espinosa, L. E., ... & Federico, P. (2017). Interrater reliability of visually evaluated high frequency oscillations. Clinical Neurophysiology, 128(3), 433-441.